In molecular biology, the synthesis of RNA is a critical process with broad applications, particularly in areas such as in vitro transcription (IVT). Traditionally, wild-type T7 RNA polymerase has been the go-to enzyme for this purpose, thanks to its ability to efficiently catalyze the 5'→3' synthesis of RNA from double-stranded DNA templates. However, a common challenge with this approach has been the unwanted production of double-stranded RNA (dsRNA) by-products, which can interfere with downstream applications and reduce the overall quality of RNA.
Addressing the dsRNA Challenge
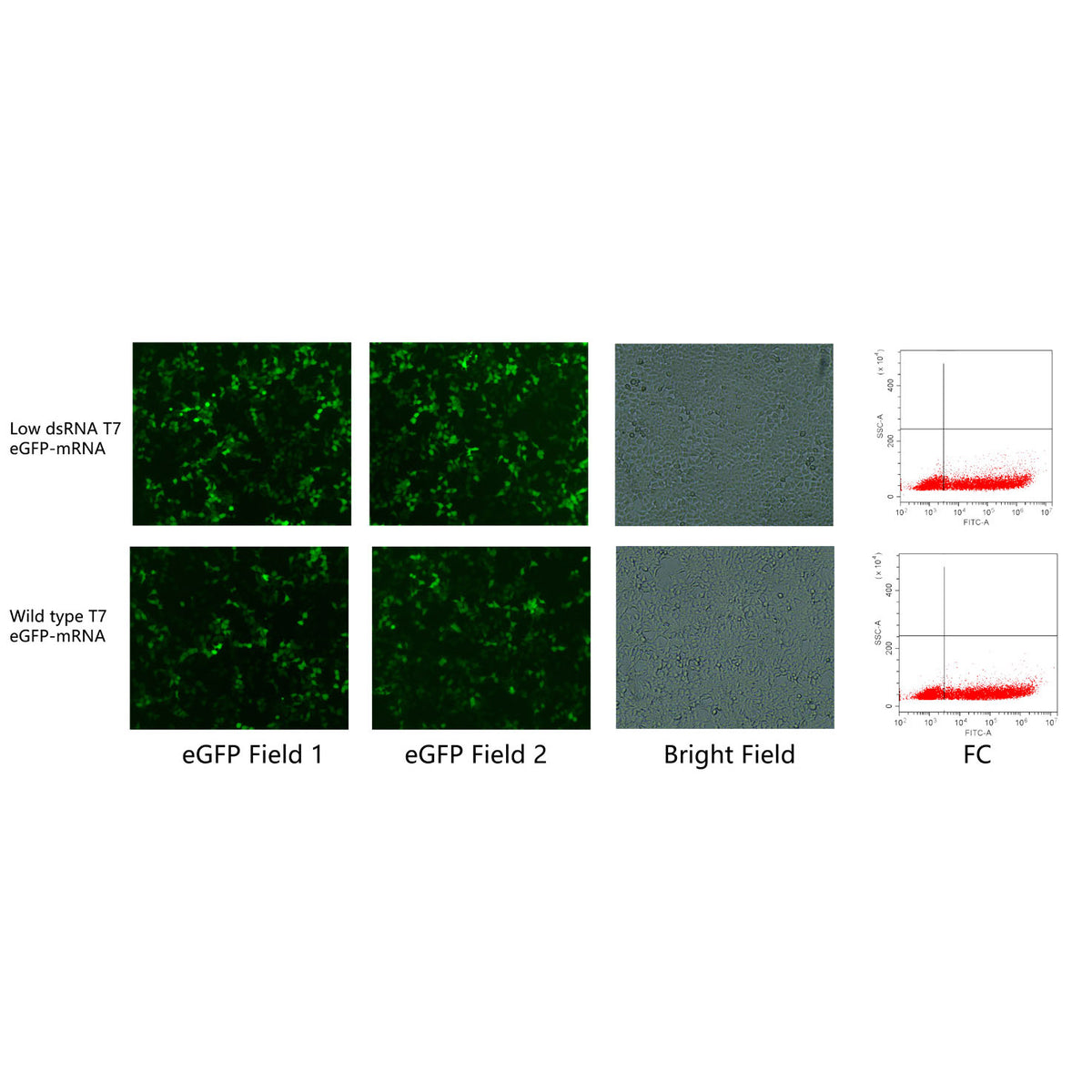
Recent advancements at Yeasen Recent advancements have led to the development of engineered variants of T7 RNA polymerase that significantly reduce dsRNA production while maintaining high transcription efficiency. This innovation is particularly valuable for researchers who require high-purity RNA with minimal contaminants. The reduction in dsRNA by-products ensures that the RNA synthesized is more suitable for sensitive applications, such as therapeutic RNA production, where purity is paramount.
Compatibility and Efficiency
One of the notable features of these engineered RNA polymerases is their compatibility with cap analogs, such as Trilink CleanCap AG. This compatibility is essential for producing capped RNA, which is often required for translation in eukaryotic systems. Despite the modifications to reduce dsRNA levels, these polymerases maintain high yields, comparable to the wild-type enzyme, making them a reliable choice for researchers who need both efficiency and purity in their RNA synthesis.
Implications for Research and Development
The ability to synthesize RNA with lower dsRNA content has broad implications for both basic research and clinical applications. For example, in vaccine development or gene therapy, where RNA is a key component, the reduction of dsRNA can lead to fewer off-target effects and better overall performance of the therapeutic RNA. This advancement not only improves the quality of the synthesized RNA but also enhances the safety and effectiveness of RNA-based technologies.
Future Perspectives
As molecular biology continues to advance, the demand for high-quality RNA synthesis will only grow. Innovations that address common challenges, such as dsRNA by-product reduction, are paving the way for more efficient and effective research and therapeutic development. By optimizing RNA polymerase variants, researchers can achieve better results in less time, contributing to the rapid progress of the field.
CleaScrip™ T7 RNA Polymerase (low dsRNA, 250 U/μL)
This is an engineered T7 RNA polymerase variant (low dsRNA) derived from the wild-type T7 RNA polymerase and produced in Escherichia coli. It significantly reduces the production of double-stranded RNA (dsRNA) while efficiently incorporating cap analogs, and exhibiting highly efficient in
CleaScrip™ T7 RNA Polymerase (low dsRNA, 250 U/μL)
vitro transcription (IVT) comparable to the wild-type T7 RNA polymerase. It catalyzes the 5'→3' synthesis of RNA on double-stranded DNA from its T7 promoter sequence (5'-TAATACGACTCACTATAG*-3') and uses NTPs as substrates